More detailed help pages are available at http://www.blaststation.com/help/bslocal/en/mac/index.html
or Launch BlastStation-Local and goto Help > BlastStation Help Menu.
Double click
BlastStation-Local icon, which is in the Applications folder.
 |
Before performing local BLAST search, you have to download database files from the NCBI ftp server or create database files from FASTA
or FASTQ data.
Downloading Databases
If you always want to perform local BLAST searches against most recent NCBI
databases, Synchronized Database Download function in
BlastStation-Workgroup
is suitable. BlastStation-Workgroup
can perform synchronized download of any FASTA data
on the Internet and create local databases automatically, too.
On the Tools menu, click Download DB. The DB Download window opens. DB Type column indicates that the database file is in DNA or Protein database format. For example env_nr.tar.gz is used for blastp and env_nt.tar.gz is used for blastn.
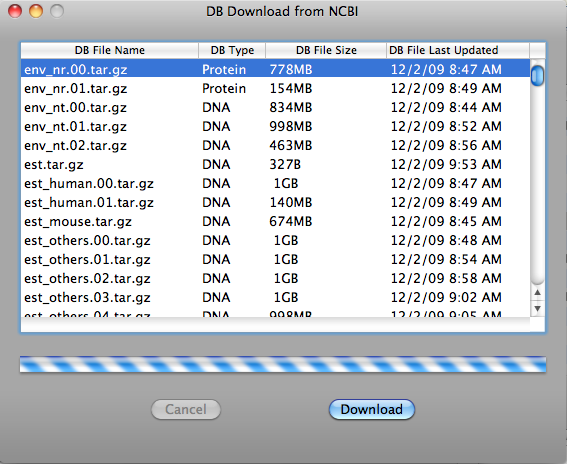
Click database file to highlight and then click "Download" button to download it. If you want to cancel download, click "Cancel" button during downloading.
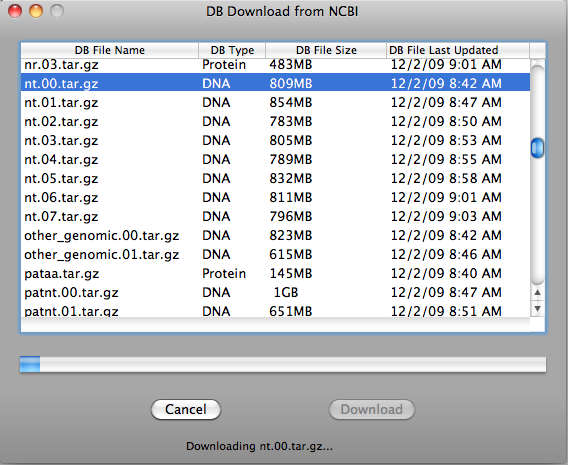
When download is finished, database file will be automatically placed in the database directory and ready to use.
Creating your own Databases
You can create database files from your own multi-FASTA data
or FASTQ data. Plain, .gz, .Z, and .zip text files are accepted.
FASTA data format is defined here(http://www.ncbi.nlm.nih.gov/blast/html/search.html) and multi-FASTA data format is defined here(http://baboon.math.berkeley.edu/mavid/multifasta.html).
On the Tools menu, click Create DB. The Create DB window opens.
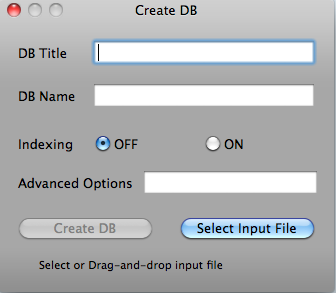
Clicking "Select Input File" button will open File Open dialog. Select input file and click "Open" button.
Or drag and drop input file.
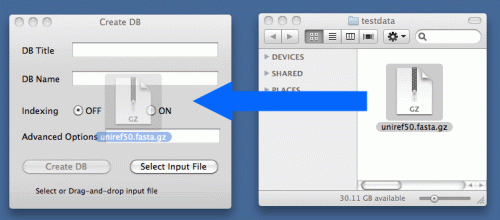
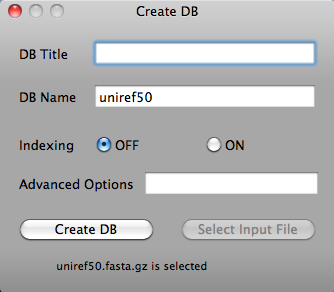
Clicking "Create DB" button creates database. When database is created, "Database is created" message will be shown. The other options are for advanced users. Please refer to http://www.blaststation.com/help/bslocal/en/mac/Chap2/02_CreateDB.html for details.
BlastStation-Local64 Only
When selected input file is large, multiple database files such as nt.00, nt.01, and so on will be created. At the same time, the alias file, such as nt.nal will be created. When this alias file is selected as database, Blast+ search over large virtual database can be done.
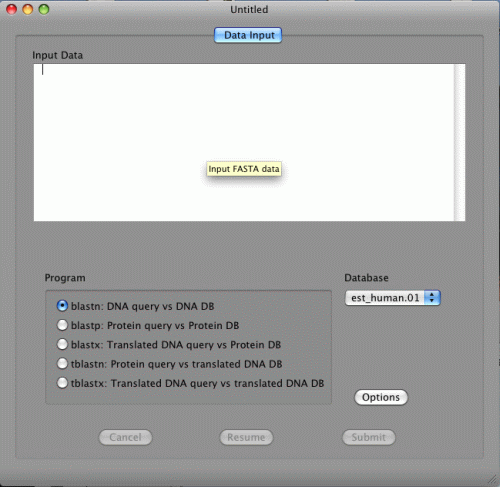
Start BLAST Search
Copy and paste FASTA data below in the Input Data.
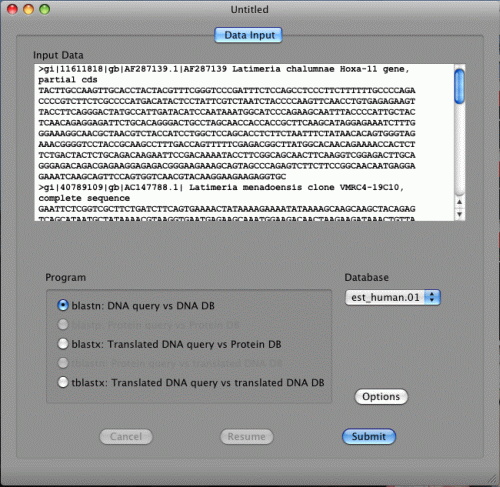
blastn, blastx, and tblastx are available in the Program section.
This is because the FASTA data are DNA sequences. If the data are protein sequences, blastp and tblastn will be available.
If blastn is selected, DNA databases will be shown in the Database pull down menu. If blastp is selected, protein databases will be shown on the other hand. Two databases, NucleotideExample and ProteinExample, are installed with BlastStation-Local. You can use them for testing.
If you have a FASTA file on your Mac, you can also drag and drop it to the Input Data area.
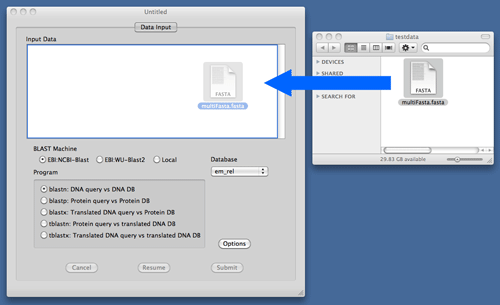
BlastStation-Local64 Only
BlastStation-Local64 can select alias files such as .nal or .pal which ties up multiple database file. Alias files will be made through Create DB or included downloaded database file.
Just click blue "submit" button. The following dialog will be shown.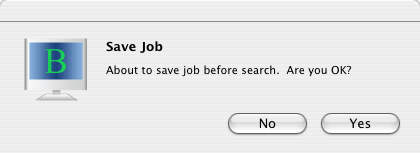
Click "Yes".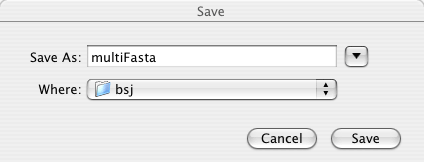
Enter file name and click "Save". This file name will be the BLAST search job name.
That's it! Now BLAST search is started. The status bar is showing "Searching...".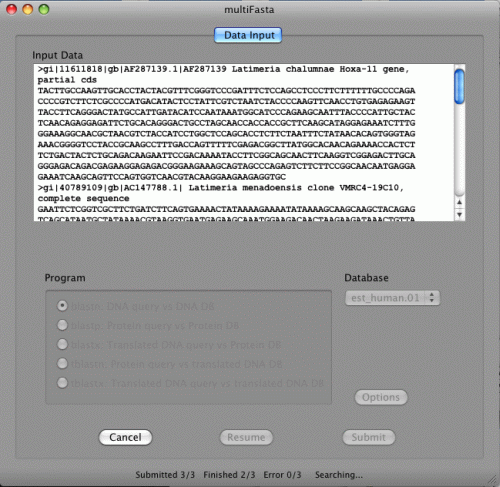
When BLAST search is finished, the following dialog will be displayed.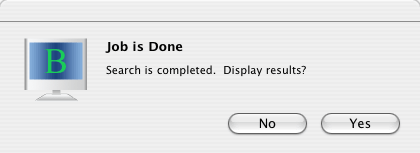
Click "Yes" to see search results.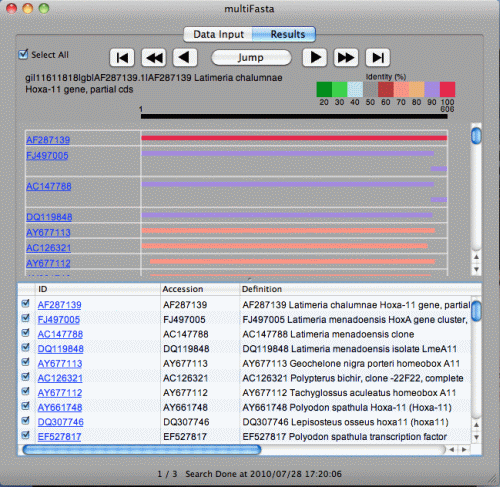
Bar graph is shown on the upper half. The color of the bar is classified by the identity. Red is 100%, purple is 90% or more, and so on. The length of the query sequence (606 in this case) is given above the black bar at the top. You can easily understand which portion of the subject sequence matches with the query sequence. Move your mouse cursor on the bars. The color of the bar changes and four numbers will be displayed.
Table is shown on the lower half, which consists of ID, Accession number, Definition, length, bit score, E Value, and Identity.
In order to display the other search results, click arrows below. Previous Result
Previous Result Next Result
Next Result Fast Backward
Fast Backward Fast Forward
Fast Forward First Result
First Result Last Result
Last Result
When you click "Jump" button, the drawer will be shown next to the window. Description of each FASTA file will be listed in this drawer. Clicking each description will display corresponding search result. If you have many descriptions, type in search strings in the text field and hit enter to narrow this list. Clicking "Jump" button again will hide the drawer.
Search results can be exported as an Excel readable CSV file from File > Export menu. There are "All Results" and "All Best Hits" options.

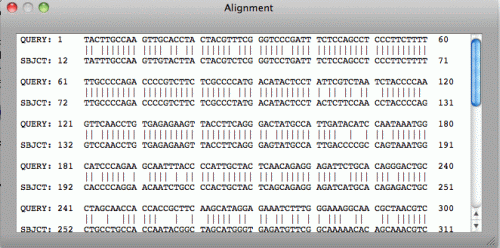
Job > Display Alignment menu will open Alignment window. The detailed alignment
information will be displayed when the bar graph in the Results pane is clicked.
Job > Display Results menu will display Blast results file in the default browser.

Check check boxes of sequences to be exported. "Select All" will check all sequences. "Select All" again will uncheck all sequences.
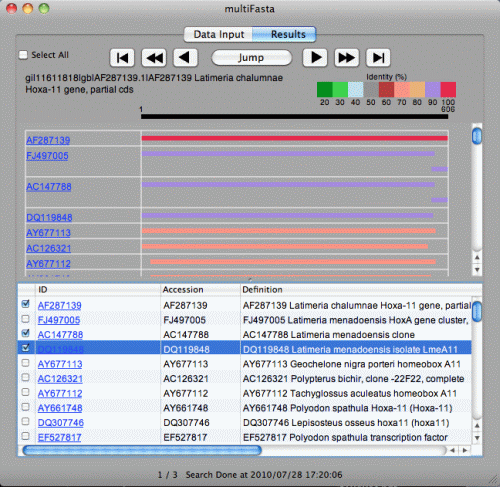
click File > Export FASTA menu. The window below will be shown.
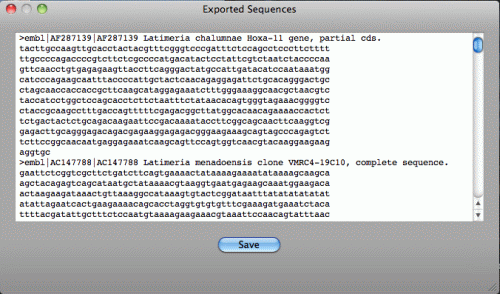
This FASTA data will be saved in the file by clicking "Save".
More detailed help pages are available at http://www.blaststation.com/help/bslocal/en/mac/index.html
or Launch BlastStation-Local and goto Help > BlastStation Help Menu.
Copyright © 2005 - 2011 TM Software, Inc. All rights reserved.